Discovery of tuberculosis bacterium enzyme paves way for new drugs
22 April 2009
A team of University of Maryland scientists has paved the way for the development of new drug therapies to combat active and asymptomatic (latent) tuberculosis infections by characterizing the unique structure and mechanism of an enzyme in M. tuberculosis, the bacterium that causes the disease.
Assistant Professor of Chemistry and Biochemistry Barbara Gerratana, in the university's College of Chemical and Life Sciences, led the research team, which included her graduate student Melissa Resto and Assistant Professor Nicole LaRonde-LeBlanc.
"The NAD+ synthetase enzyme that our study describes is absolutely essential for the survival of the tuberculosis bacteria and an important drug target. We can now use the information we have about its structure and mechanism to develop inhibitors for this enzyme," Gerratana explained.
The study, titled 'Regulation of active site coupling in glutamine-dependent NAD+ synthetase',' was published on March 8, 2009 in Nature Structural & Molecular Biology.
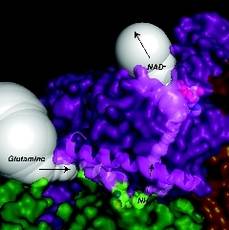
Close up of the glutamine and ammonia tunnels shown
in white surface connecting the glutaminase and synthetase active sites
from two different subunits. DON bound in the glutaminase active site of
the green subunit +and NaAD +bound in the synthetase active site of the
magenta are shown in CPK.
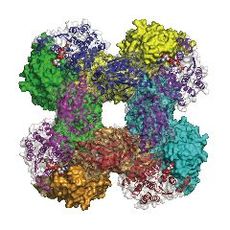
Quaternary structure of NAD+ synthetaseGln.
Glutaminase domains line the central cavity, whereas synthetase domains
are on the outside. Glutaminase domains in the top ring are illustrated
in ribbon with a transparent surface representation. NaAD+ bound in the
synthetase active site is shown in CPK.
The development of new drugs to combat tuberculosis (TB) has become urgent, as strains of TB resistant to all major anti-TB drugs have emerged worldwide. The World Health Organization estimates that one third of the world's population is asymptomatically infected with TB and that ten percent will eventually develop the disease.
According to other leading TB researchers, these new findings from Gerratana and her colleagues will be extremely valuable for the design of structure-based inhibitors specific for M. tuberculosis NAD+ synthetase and may lead to the development of new drugs to combat and eliminate the disease.
"NadE [NAD+ synthetase] represents one of a small handful of TB drug targets that has iron-clad validation, the lack of a crystal structure was the only serious impediment to drug development and this study represents a hugely important step forward" said Clifton E. Barry, Chief of the Tuberculosis Research Section of the Intramural Research Division of the National Institute of Allergy and Infectious Diseases.
"Inhibiting NadE even kills non-replicating cells, so this discovery may well benefit the one-third of the human population that carries latent bacteria."
NAD+ synthetase is responsible for making NAD+, a coenzyme found in all living cells that is involved in regulating many cellular processes and in reduction-oxidation metabolic reactions.
More than one biosynthetic pathway is usually involved in NAD+ production. In humans, NAD+ can be obtained through several different complex pathways, and not all of the pathways utilize NAD+ synthetase to produce NAD+.
Unlike in humans, however, there are only two pathways involved in producing NAD+ in the tuberculosis bacterium and both depend on the activity of NAD+ synthetase to obtain NAD+.
"We are optimistic about the potential for developing new drugs that will effectively target this enzyme in TB and minimize side effects to humans, since we have NAD+ biosynthetic pathways that are independent of the NAD+ synthetase activity," Gerratana said.
The World Health Organization reports that a new instance of TB infection occurs every second. Current treatment of tuberculosis targets the active tuberculosis bacterium and has little effect on the non-replicating bacterium. "If we don't tackle latent tuberculosis, this disease will not be eradicated," Gerratana said.
Bookmark this paget>