Common weak point in flu virus strains could lead to universal flu vaccine
6 March 2009
Two recent studies have found a common Achilles’ heel in a wide range of seasonal and pandemic influenza A virus strains.
Researchers at The Scripps Research Institute in La Jolla, California, have found an infection-fighting human protein, or human antibody, that neutralizes various influenza A virus subtypes by attaching to these viruses in the same place.
The study provides data about the antibody attachment site that are similar to the findings of another research group led by Dr Wayne Marasco, associate professor of medicine at the Dana-Farber Cancer Institute and Harvard Medical School in Boston. This group identified a small family of lab-made proteins that neutralize a broad range of influenza A viruses, including the H5N1 avian virus, the 1918 pandemic influenza virus and seasonal H1N1 flu viruses.
These human monoclonal antibodies, identical infection-fighting proteins derived from the same cell lineage, also were found to protect mice from illness caused by H5N1 and other influenza A viruses. Because large quantities of monoclonal antibodies can be made relatively quickly, after more testing, these influenza-specific monoclonal antibodies potentially could be used in combination with antiviral drugs to prevent or treat the flu during an influenza outbreak or pandemic.
This common attachment site provides a constant region of the flu virus for scientists to target in an effort to develop a so-called universal flu vaccine. Such a vaccine would overcome the annual struggle to make the seasonal flu vaccine match next year’s circulating flu strains and might help blunt emerging pandemic influenza viruses as well.
The Scripps research team, led by Dr Ian A Wilson, in collaboration with researchers at the biopharmaceutical company Crucell Holland (The Netherlands), discovered the potent antibody during a systematic examination of blood samples taken from healthy individuals who previously had been vaccinated with the ordinary seasonal flu vaccine. The results have been published in the journal Science [1].
Using sophisticated screening technologies, the team isolated antibodies that recognize flu viruses to which the average person has never been exposed, such as H5N1 avian flu viruses.
Through this process, the scientists found one antibody called CR6261 that had broad neutralizing capabilities. Subsequently, they found several antibodies similar to CR6261 in other donors as well. With the help of a robotic crystallization laboratory, the Scripps team quickly determined the detailed three-dimensional structures of this antibody when bound to the H1 virus that caused the 1918 pandemic flu as well as to an H5 virus with pandemic potential.
CR6261 bound to a relatively hidden part in the stem below the mushroom-shaped head of the hemagglutinin protein, one of two major surface proteins found on the flu virus.
In the second study [2], Dr Marasco and his colleagues discovered and described the atomic structure of an obscure but genetically stable region of the influenza virus to which their monoclonal antibodies bind. The hidden part of the influenza virus is in the neck below the peanut-shaped head of the hemagglutinin (HA) protein. HA and neuraminidase are the two main surface proteins on the influenza virus.
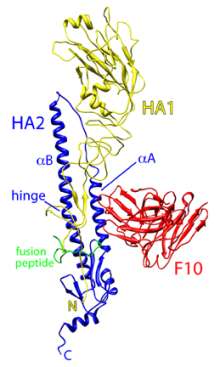 |
| Ribbon diagram of the influenza virus H5 hemagglutinin (HA) surface protein bound by the F10 monoclonal antibody (red). The two chains of H5 are HA1 (yellow) and HA2 (blue). (Credit: William Hwang and Jianhu Su, Dana-Farber Cancer Institute) |
Dr Marasco, Jianhua Sui, MD, PhD, and other Dana-Farber colleagues began their study with avian flu viruses. They scanned tens of billions of monoclonal antibodies produced in bacterial viruses, or bacteriophages, and found 10 antibodies active against the four major strains of H5N1 avian influenza viruses.
Encouraged by these findings, they collaborated with Ruben O Donis, PhD, of the US Centers for Disease Control (CDC) Influenza Division, and found that three of these monoclonal antibodies had broader neutralization capabilities when tested in cell cultures and in mice against representative strains of other known influenza A viruses.
Influenza A viruses can include any one of the 16 known subtypes of HA proteins, which fall into two groups, Group 1 and Group 2. Their monoclonal antibodies neutralized all testable viruses containing the 10 Group 1 HAs — which include the seasonal H1 viruses, the H1 virus that caused the 1918 pandemic and the highly pathogenic avian H5 subtypes — but none of the viruses containing the six Group 2 HAs.
Simultaneously, Dr Marasco’s group teamed up with Robert C. Liddington, PhD, professor and chair of the Infectious and Inflammatory Disease Center at Burnham, to determine the atomic structure of one of their monoclonal antibodies bound to the H5N1 HA. Their detailed picture shows one arm of the antibody inserted into a genetically stable pocket in the neck of the HA protein, an interaction that blocks the shape change required for membrane fusion and virus entry into the cell.
When they surveyed more than 6,000 available HA genetic sequences of the 16 HA subtypes, they found the pockets to be very similar within each Group but to be significantly different between the two Groups. The genetically stable pockets, they note, may be a result of evolutionary constraints that enable virus-cell fusion. This could also explain why they did not detect so-called escape mutants, viruses that elude the monoclonal antibodies through genetic mutation.
“One of the most remarkable findings of our work is that we identified a highly conserved region in the neck of the influenza hemagglutinin protein to which humans rarely make antibodies,” says Dr. Marasco. “We believe this is because the head of the hemagglutinin protein acts as a decoy by constantly undergoing mutation and thereby attracting the immune system to produce antibodies against it rather than against the pocket in the neck of the protein.”
Their findings could also assist vaccine developers. Current influenza vaccines target the constantly mutating head of the HA protein and do not readily generate antibodies against the conserved region in the neck.
“An important goal is to redirect the immune response of vaccines to this invariable region of the hemagglutinin to try to obtain durable lifelong immunity,” Dr Marasco states. The monoclonal antibodies identified in their paper are very well-characterized, Dr Marasco notes, and he is optimistic about their further clinical development.
“These are fully human monoclonal antibodies that are ready for advanced preclinical testing,” he says. He currently is arranging to use NIAID research resources to take the next steps: first, testing the antibodies in ferrets, the gold standard animal model for influenza, and then developing a clinical grade version of one antibody that could enter human clinical trials as soon as 18 months from when the development program begins. Should the antibodies prove safe and effective in humans, it could take several years to develop a licensed product.
Emergency planning still required
Dr Jean Challiner, Medical Director of Clinical Solutions, a UK supplier of clinical decision-support systems, says that it is still essential to maintain disease-outbreak response systems: “It is of course welcome news indeed that new treatments are being developed to protect the population from the devastating consequences Pandemic Influenza would currently have. However, development of effective antiviral medicine alone is not the only requirement to both prepare for and manage the consequences of an outbreak, and we must not become oblivious to the threats posed by other diseases, such as SARS — for which there is no specific treatment or vaccine available.
“The outbreak of any pandemic can be manageable if effective measures are taken to prepare for outbreaks in advance — and the development of antiviral medication, although crucial, only makes up part of this. Offering the public access to healthcare professionals who can carry out an appropriate assessment and offer up-to-date advice and information, will reduce public anxiety, support effective use and distribution of medicines available, and help contain the outbreak so minimising risk.
"Having the right tools to support healthcare professionals involved, as well as the ability to collect and analyse information gathered, is an essential part of forward planning — and this is where technology can lend a hand.
“In an emergency situation traditional access to healthcare is not always an option. Technology can deliver web and telephony-based support for assessment, direction of care, advice, guidance and, where appropriate, authorisation of antiviral medicine treatment without the need for people to leave their homes.
"Technology can also be used in a command centre environment to help monitor the situation in real-time with the facility to rapidly change and update the instructions available on websites and for call operators who are communicating with the public.
“The onset of Pandemic Influenza is well overdue and we cannot be sure when the next pandemic will strike. For that reason it is essential that we are prepared as a nation to effectively manage a pandemic threat through provision of systems which will give the general public easy access to consistent assessment processes and advice — so ensuring the most effective implementation of immunisation and treatment regimes and maximising the benefit that new medicines can provide.”
References
1. DC Ekiert et al. Antibody recognition of a highly conserved influenza virus epitope. Science. DOI 10.1126/science.1171491 (2009).
2. J Sui et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Structural & Molecular Biology. DOI: 10.1038/nsmb.1566 (2009).
Further information
The Dana-Faber Cancer Institute website has an animation of human monoclonal antibodies and the flu virus.
For more information on influenza see the US National Institute of Allergy and Infectious Diseases's (NIAID) flu portal and the Centers for Disease Control's Seasonal Flu page.
Also visit www.PandemicFlu.gov for a one-stop access to US Government information on avian and pandemic flu.
Bookmark this page/font>