| Diagnostic imaging | |||
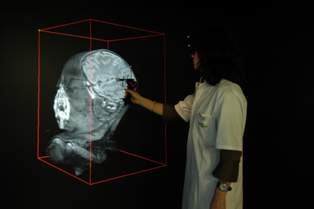
Erasmus University Medical Centre develops interactive immersive 3-D images25 April 2006 Rotterdam, Netherlands & Mountain View California. Erasmus University Medical Centre (Erasmus MC) in Rotterdam has developed 3D volume rendering software, capable of converting 2D medical images to 3D. The software runs in an immersive, interactive environment called I-space, developed at the Center. It presents clinicians with new ways to investigate datasets from all kinds of 3D imagery, including MRI and CT scans.
For the commercial launch of the I-space model, the medical centre selected Barco projectors and Silicon Graphics Prism visualization systems as image generators to create the four-sided, 3x3x3 metre 3D world intended for multidisciplinary collaboration. Because the prototype model at Erasmus MC surpassed even the highest expectations of researchers and clinicians, the medical centre spun off a Rotterdam-based company, Crosslinks, which is now beginning to market I-space to hospitals and medical centres throughout Europe in collaboration with SGI and Barco. The original I-space at Erasmus MC was developed in close cooperation with medical staff that use the technology regularly for complex diagnoses. While I-space, in theory, could be used for various virtual reality applications the proprietary software is specifically targeted to the medical and biology fields. "We chose the Silicon Graphics system for I-space because we needed hardware that was both suitable for high-performance computing and also delivered superb graphical capabilities," said Ronald Nanninga, founder and managing director of Crosslinks. "There are eight graphics pipes in the system — you are actually standing inside the data — so we required a visualization computer that is powerful enough to really do the rendering of the 3D software in a very efficient way, and only SGI had the appropriate solution. When we began working on the development of I-space, we used an older SGI system, but with this brand new Prism, our performance really improved. Frame rate has gone up from 4 to 15, and that tells you something about how easy it is to move around large data sets, turn them around, and zoom in and zoom out. It's easier to make a diagnosis together with other medical personnel when the surgical reality is right in front of you, rather than seeing it alone, just on a small computer screen." A Silicon Graphics Prism visualization system with 8 Intel Itanium 2 processors, 8 ATI FireGL graphics processors and 12GB of memory is used to drive I-space. I-space enables researchers to explore vast amounts of genomics and proteomics data in an infinite 3D world. It also presents clinicians with new ways to investigate datasets from all kinds of 3D imaging modalities, including MRI and CT scans. I-space uses 8 Barco projectors for the four walls: the floor, and the left, right and front. A 3D mouse uses four tracking devices — one in each corner, enabling the system to recognize the relative position of the mouse — and a virtual stick, which allows the user to touch an object, push it on one side, zoom in/zoom out, and even slice the object. Stereoscopic glasses complete the immersive 3D experience. I-space was specifically created to allow doctors and researchers to discuss — among different disciplines — the data they are all seeing in the immersive visualization. "We built I-space ourselves in collaboration with the folks from SGI and Barco," said Prof. Dr. Peter van der Spek, a geneticist by training who is also an engineer. "Barco has the projectors, but the Silicon Graphics Prism has all the graphics pipes that superbly work together with Barco. It's cutting-edge technology and the power of Intel inside the Prism system is also very important for us, so we are very, very happy with the technology that drives I-space." I-space at Erasmus Medical Center At Erasmus MC, I-space is used for two reasons: research and clinical diagnostics. The clinical application deals with medical visualization in 3D of different modalities such as MRI scans, CT scans, and ultrasound images. Doctors can walk through an MRI scan of a patient while discussing it with their colleagues. Multidisciplinary discussions are routine. For instance, for a patient with a brain tumour, the neurologist, with the neurosurgeon, are together in I-space and can decide what the best strategy is to remove the tumour from the brain. Or, for a person with a lung tumour, the thorax surgeon is there with the lung specialist, and they look to see where the tumour is positioned. Is it an area accessible to the thorax surgeon, and how can he most optimally approach the tumour? Are there large blood vessels going through the tumour? Is the tumor close to the heart? All of this has to be taken into account when removing a tumour. Additional clinical applications
"We use the I-space for very different reasons," said Prof. Prof. Dr. van der Spek. "We render the images, scan the images, into the I-space with the Silicon Graphics computer. We stack the images on top of each other and then we project them. We can do all kinds of very sharp visualizations, which in turn allow us to do very precise measurements on the MRI scans, on CT scans or on ultrasound images. We can zoom in really close, within the scan, to make it larger, to make it more obvious where to look for the clinician. This is a very multidisciplinary effort where we open up the IT box for the doctor and his colleagues, because we store the patients' images for the doctors and process and serve them. The very strong graphics capacity of the SGI equipment, including its software and the OpenGL graphics libraries, allows us to visualize, really, life in 3D." Translational Medicine The research application of I-space with the Silicon Graphics system deals with genomics data mining and proteomics data mining for translational medicine. At Erasmus MC, a 32-processor SGI Origin 3800 server runs an Oracle database with clinical and molecular data. The configuration allows the researchers to actually link and integrate the data as well as visualize it in a very efficient manner. Based on the now-occurring convergence of molecular imaging and genomics, translational medicine couples knowledge from patients' data with other patients, in a vast database, aimed at personalized treatment of diseases. The two different fields of molecular imaging and genomics require the imaging of the patient scan with the molecular aspects, coupled with the use of genomic strategies, to come up with to novel biomarkers. Biomarkers recognize, in a very specific way, the type of tumour. Researchers around the world are dedicated to finding biomarkers that are unique for certain types of cancers. For example, if a patient has a Type A biomarker for a tumour, doctors prescribe certain medication and a specific treatment. But if the tumour is a Type B biomarker, a completely different intervention procedure would be prescribed. The benefits are tremendous, as revealed in a medical paper published by Erasmus MC in December 2005 describing the genomics application of studies of a certain type of slow-growing brain tumours (oligodendroglioma). By using a genomic strategy, Prof. Dr. van der Spek reports that doctors can decide up-front whether a patient will be chemo-sensitive or chemo-resistant. "Chemotherapy is an absolutely awful treatment and if, in advance, you can decide whether your patient will or will not respond to the treatment, that's very important information to help you make the decision of whether you are going to prescribe chemotherapy or whether you are immediately going to push the patient towards the operating theatre," said Prof. Dr. van der Spek. "So we use the genomic strategy in combination with the medical imaging in the I-space run by the SGI computer in this new paradigm of translational medicine." "SGI technology has long been a leader in biomedical research and drug discovery. The introduction of I-space brings SGI and partners Barco and Crosslinks to the forefront of both medical research and clinical applications, opening up the amazing possibilities of multidisciplinary collaboration of doctors and surgeons for the specific treatment of individuals," said Afshad Mistri, market segment manager, Sciences, SGI. "We are at the dawn of a new era of personalized medicine, made possible by advances in science and SGI technology."
|